Collapse the tips of a phylogenetic tree based on a temporal threshold
Source:R/prune_tips.R
prune_tips.RdThis function collapses, or prune, the tips of a phylogenetic tree based on an inputted temporal threshold.
Arguments
- tree
phylo. An ultrametric phylogenetic tree in the "phylo" format.
- time
numeric or numeric vector. A numeric value, or vector, containing the temporal threshold(s) in million years for pruning the tree tips. It must be within the interval of the tips ages present in the phylogenetic tree.
- qtl
logical. A logical value indicating whether the user wants to use the quantile values of the tip ages to prune the tree. Default is FALSE
- method
numerical. A numerical value indicating the method to prune the tips. If "method = 1", the function will prune the tips originating after an inputted temporal threshold. If method = 2, the function will prune the tips originating before a temporal threshold. Default is 1.
Value
The function returns a pruned tree in the "phylo" format if a single temporal threshold was inputted. Otherwise, if a vector of thresholds was inputted, it returns an list of pruned trees.
Details
It uses the tip ages location in relation to an inputted time threshold to prune the phylogenetic tree. Setting "method = 2" makes slices based on the quantile distribution of tip ages available within the phylogenetic tree.
References
See the tutorial on how to use this function on our website.
See also
Other slicing methods: squeeze_root(),squeeze_tips(),squeeze_int(),phylo_pieces()
Author
Matheus Lima de Araujo matheusaraujolima@live.com
Examples
# Generate a random tree
tree <- ape::rcoal(20)
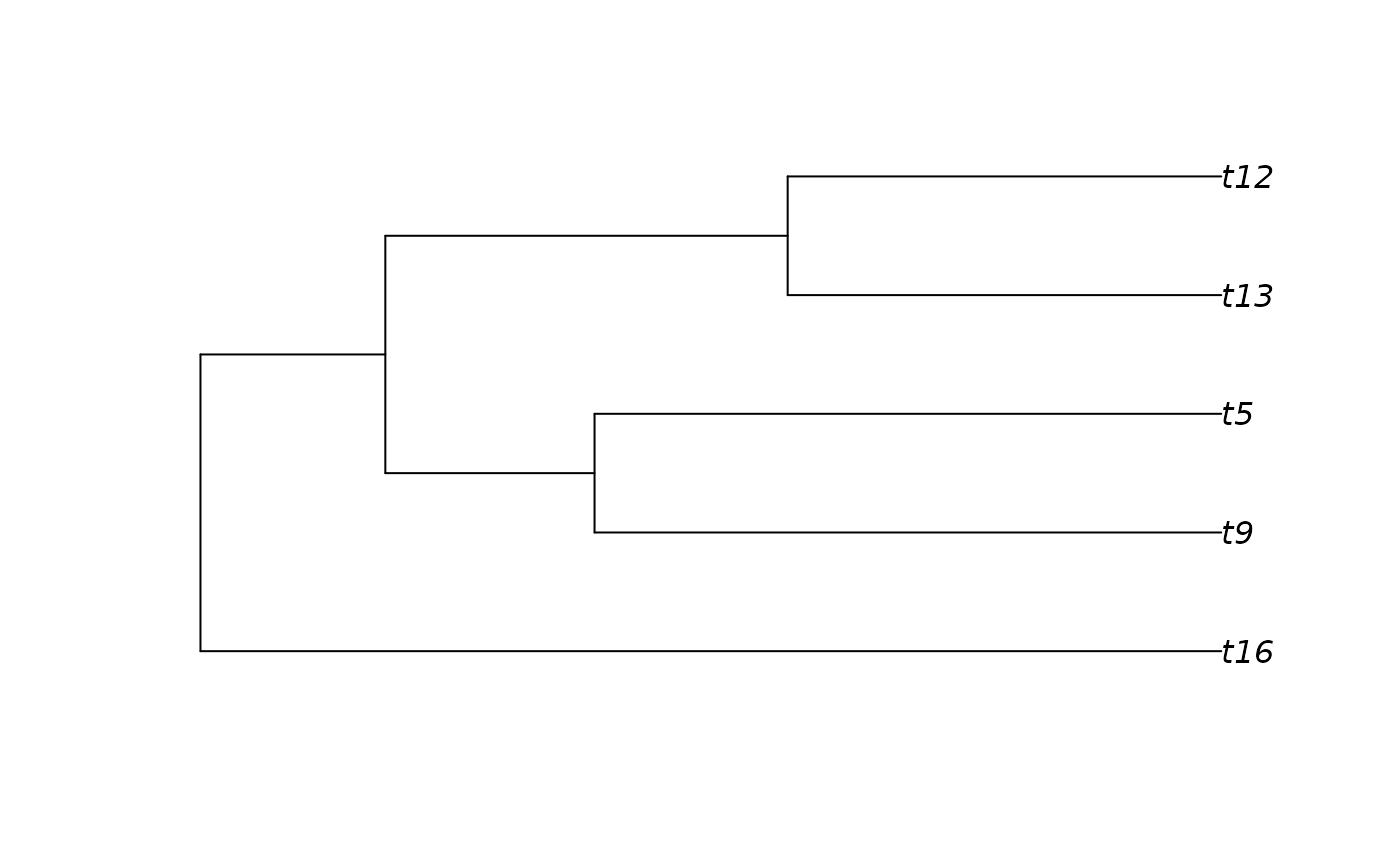
# Pruning the tips originating after 0.3 million years
tree1 <- prune_tips(tree, time = 0.1)
# Plot it
plot(tree1)
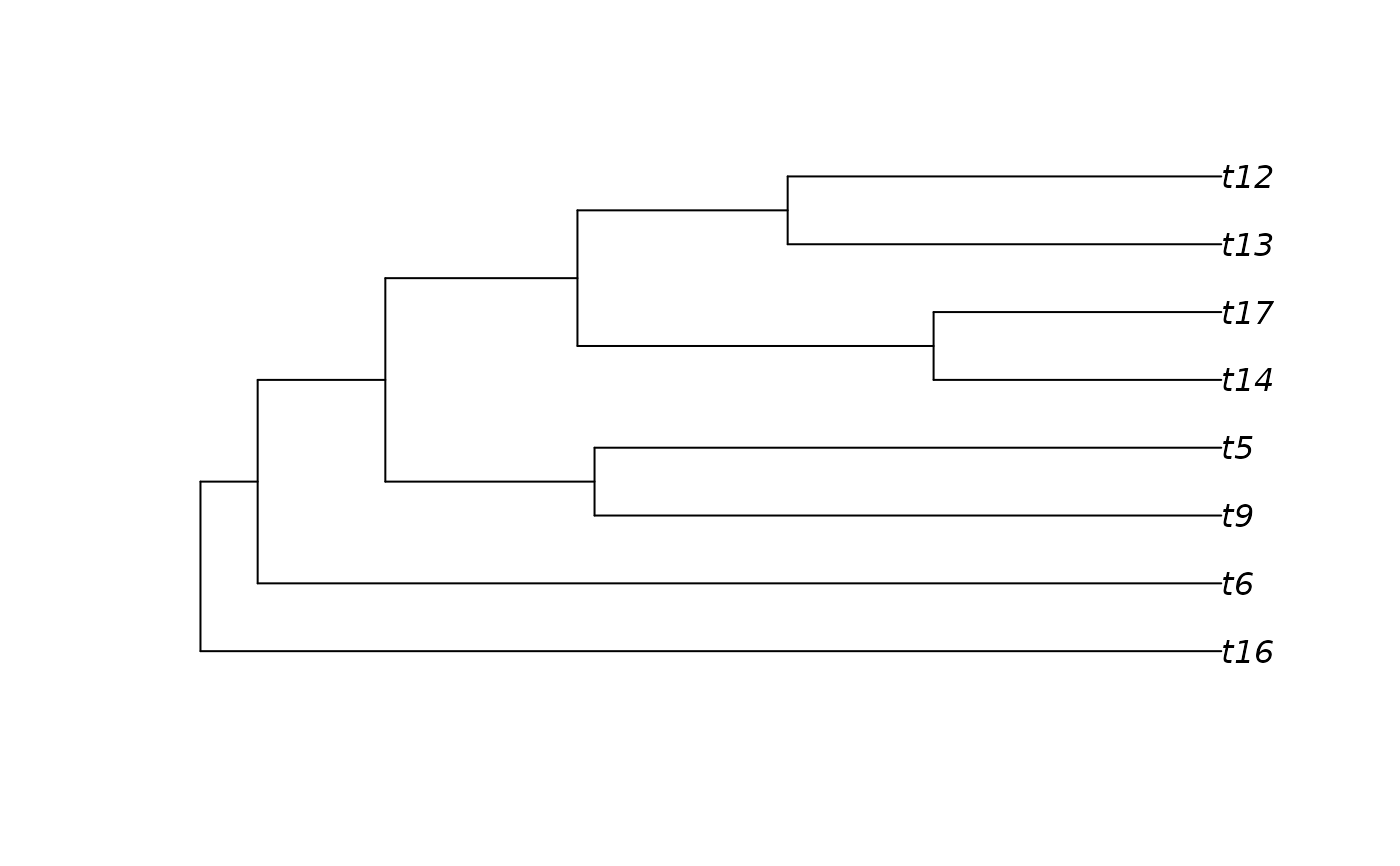 # Pruning the tips based on quantiles of tip ages
tree2 <- prune_tips(tree, time = c(0.25, 0.75), qtl = TRUE)
plot(tree2[[1]])
# Pruning the tips based on quantiles of tip ages
tree2 <- prune_tips(tree, time = c(0.25, 0.75), qtl = TRUE)
plot(tree2[[1]])
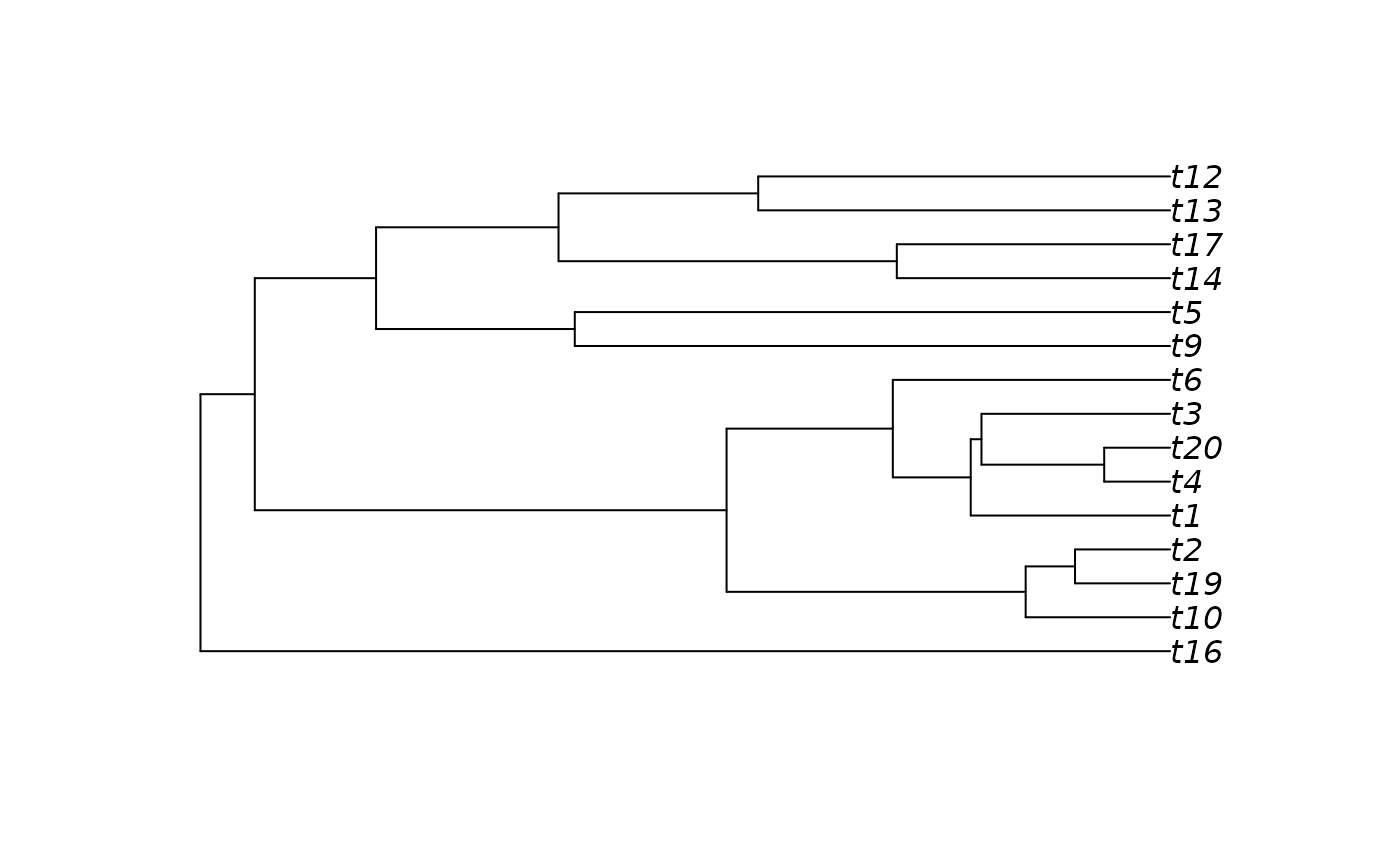 plot(tree2[[2]])
plot(tree2[[2]])