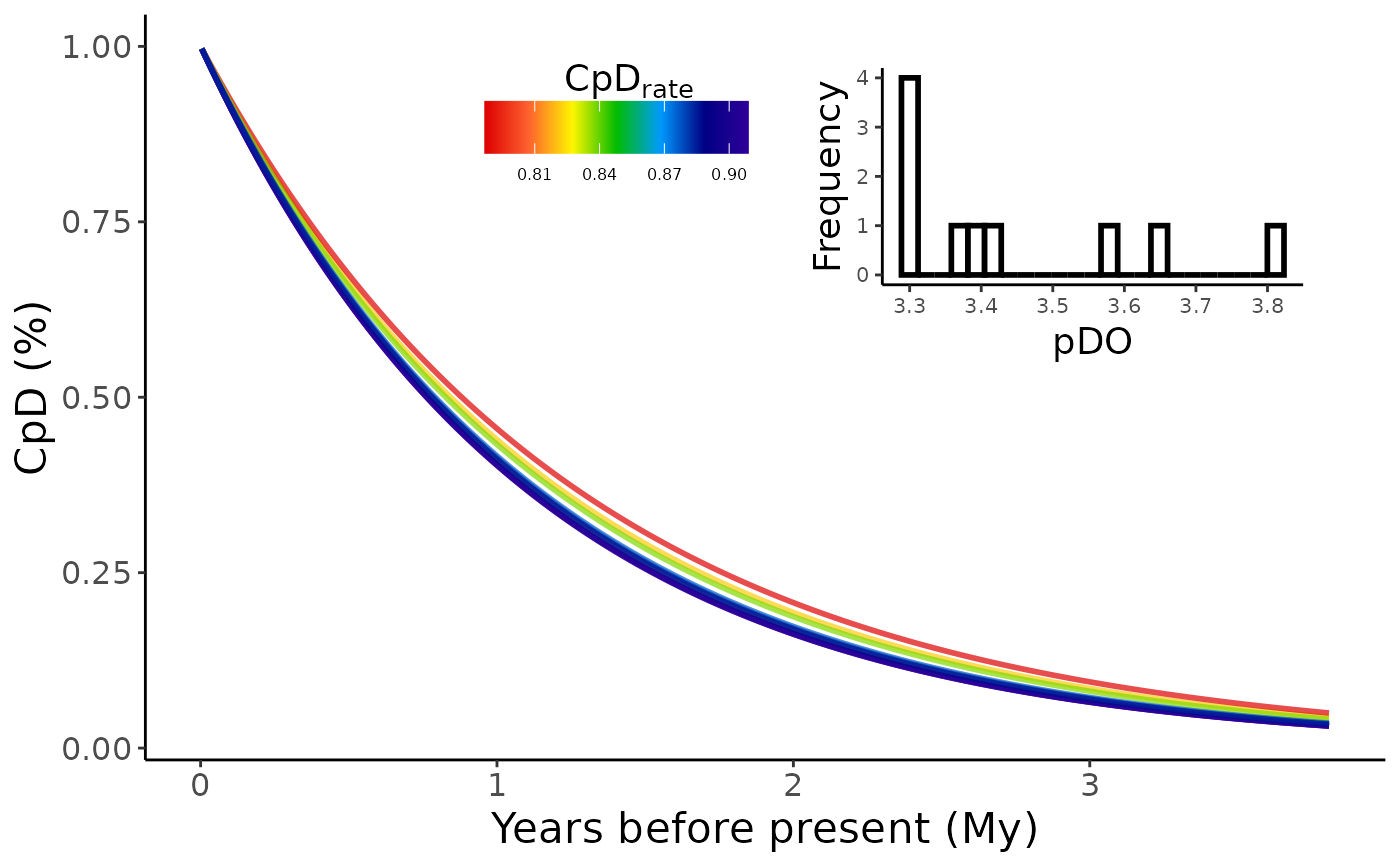
Make a line graph or a grid map of estimated rates of accumulation of a given phylogenetic index.
Source:R/CpR_graph.R
CpR_graph.RdThis function creates a line graph, or a grid map, depicting the estimated rates of accumulation of a given phylogenetic index (e.g., phylogenetic diversity, endemism, etc.), obtained from functions such as CpD(), CpE(), CpB(), or CpB_RW().
Arguments
- data
data frame. The outputted data frame from a CpR-rate function.
- rate
character string. The desired cumulative phylogenetic rate to plot, which can be phylogenetic diversity (CpD), phylogenetic endemism (CpE), phylogenetic B-diversity (CpB), or phylogenetic B-diversity range-weighted (CpB_RW). Default is NULL, but must be filled with "CpD", "CPE", "CpB", or "CpB_RW".
- map
spatial data. A grid map containing the assemblages at which the phylogenetic rates were assessed. Default is NULL.
- pal
character vector. A vector containing a color palette. If none provided, a default color palette will be used.
- qtl
boolean. Should the color palette be displayed according to CpR-rates quantiles? It can be either TRUE or FALSE. Default is FALSE.
References
See the tutorial on how to use this function on our website.
See also
Other cumulative phylogenetic rate analysis: CpD(), CpE(), CpB(), CpB_RW(), CpR_sensitivity()
Other plotting: CpR_sensitivity_plot().
Author
Matheus Lima de Araujo matheusaraujolima@live.com
Examples
# Generate a random tree
tree <- ape::rcoal(20)
# Create a presence-absence matrix
mat <- matrix(sample(c(1,0), 20*10, replace = TRUE), ncol = 20, nrow = 10)
colnames(mat) <- tree$tip.label
# Calculate the CpD for 100 tree slices
CpD_DF <- CpD(tree, n = 100, mat = mat)
#> > The 100 number of pieces inputted equals to intervals of 0.009881428781227 million of years.
# Plot it using the CpR_graph
CpR_graph(CpD_DF, rate = "CpD")
#> Warning: Ignoring unknown parameters: `size`
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the treesliceR package.
#> Please report the issue at <https://github.com/AraujoMat/treesliceR/issues>.